Imaging and Modeling of Biological Structures
Group
Angelo Cenedese – Assistant Professor at the Department of Information Engineering - University of Padova
angelo.cenedese_AT_unipd.it
Introduction
The ever-increasing availability of biological data from in vivo and in vitro experiments demands the development of new techniques and associated tools that are capable to ex- tract quantitative, specific, and relevant information of use to the researcher. Remarkably, vision based procedures play a crucial role in this framework since the visual data hold a direct link with the phenomenological (qualitative) study, in particular when morphological characteristics are in focus.
At the same time, such techniques and tools, supported by huge advances in computational resources and technology, should be endowed with a number of characteristics that allow the derivation of quantitative figures:
- rigor, that is they ought to be formal and based on clear mathematical models;
- flexibility – that is they ought to be adaptable to heterogeneous data sets in a semi-automatic manner – and applicability, that is they need to be supported by an existing software that is easily fed with the data and that directly provides the required outcome;
- computability, that is the associated software has to perform in real-time, or if off-line processing is acceptable it ought to scale well with the cardinality of the data sets.
Networked structures are among the most interesting and commonly occurring features that can be extracted from a data set [1], [2], [3], because they convey information not only on the structures per se but also on their interaction and embedding within the environment or the organism. Known examples in this sense are neuronal networks, lymphatic or blood vessels, cellular and molecular clusters, where the networked nature of these complex structures appears at different levels. Abstractly, networks are characterized by a set of local features and of a set of edges connecting different features and thus associating the local information into a global structured framework.
Thanks to the employment of time-lapse digital photog- raphy, it is often the case for biological data sets to refer to videos or image sequences, in order to study dynamic behaviors and phenomena. A software tool that is able to automatically extract features from the data and correlate them over time, would thus help the researcher to gain a quantitative understanding of the data set.
BIM-Tools
In general, given a specific biological data set, the process of information extraction can be divided in three stages, namely that of detection, tracking, and registration. Here a new unified framework is presented, for detecting, tracking, and registering network structures from dynamical live images, where the three aforementioned phases are handled in a way that is consistent both formally with a unique reference model and experimentally with the constant usage of the image data.
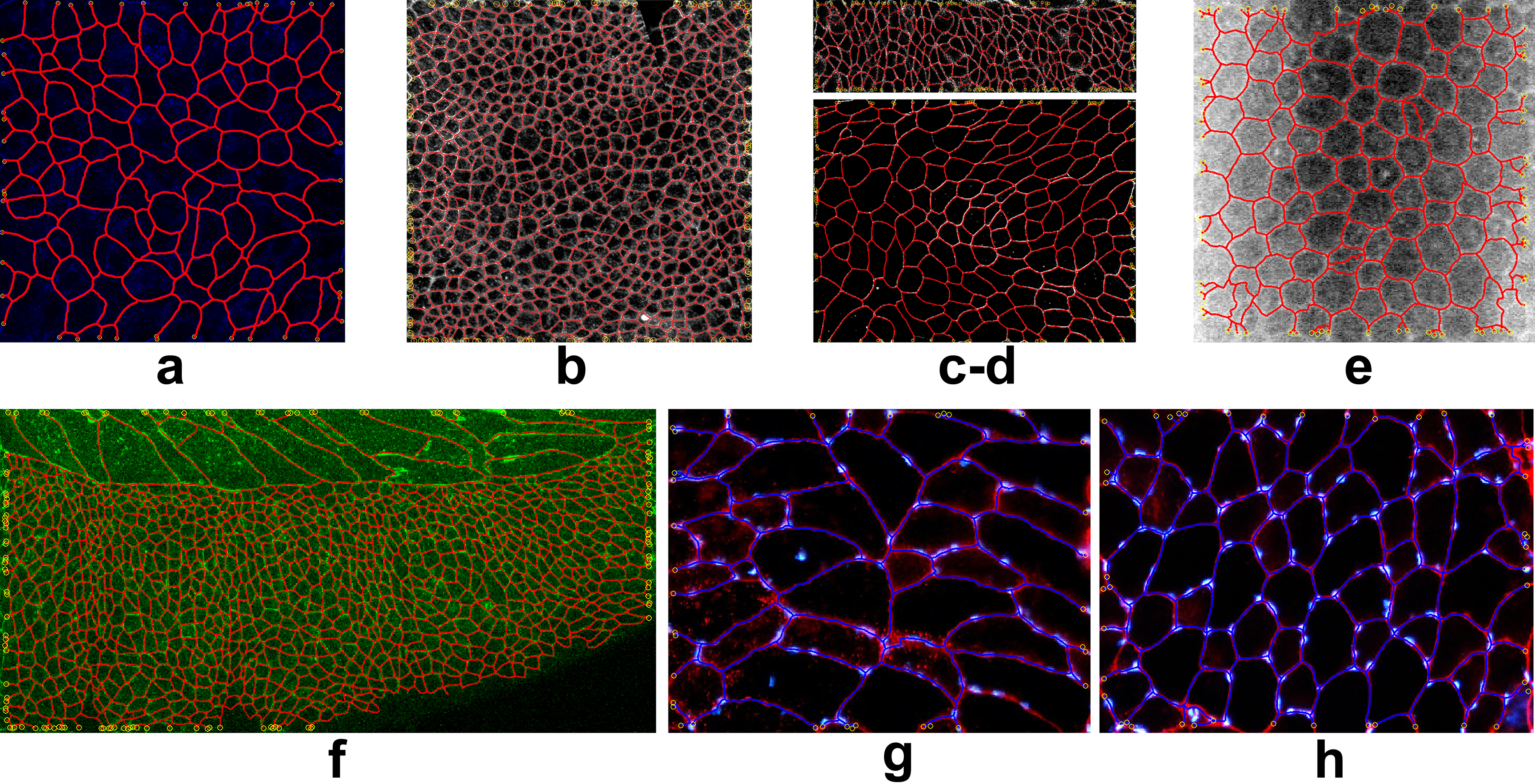
- Detection. The first available image frame is analyzed searching for an underlying network structure, which is then extracted leveraging the use of a random walk model to navigate the network edges combined with a network agent to organize the retrieved information into a complex structure. The first frame is taken as a reference frame, also for the warping function w and the outcome of this procedure is a network graph (vertices, edges) with spatial information.
- Tracking. The networked structure is then deformed in time starting from the information given by the reference frame and according to the visual data of the following frames. This phase resorts to the computation of optimal (or sub-optimal) warping maps w for motion and deformation that account for the underlying dynamics of the deformable system and at the same time accommodate the dataflow. A set of time stamped network structures is produced, each uniquely associated to the corresponding image frame.
- Registration. Once the salient structures are detected and tracked in time, is of fundamental importance to establish a quantitative relationship among them by matching networks from consecutive frames.
More details are given in the technical report.
Examples
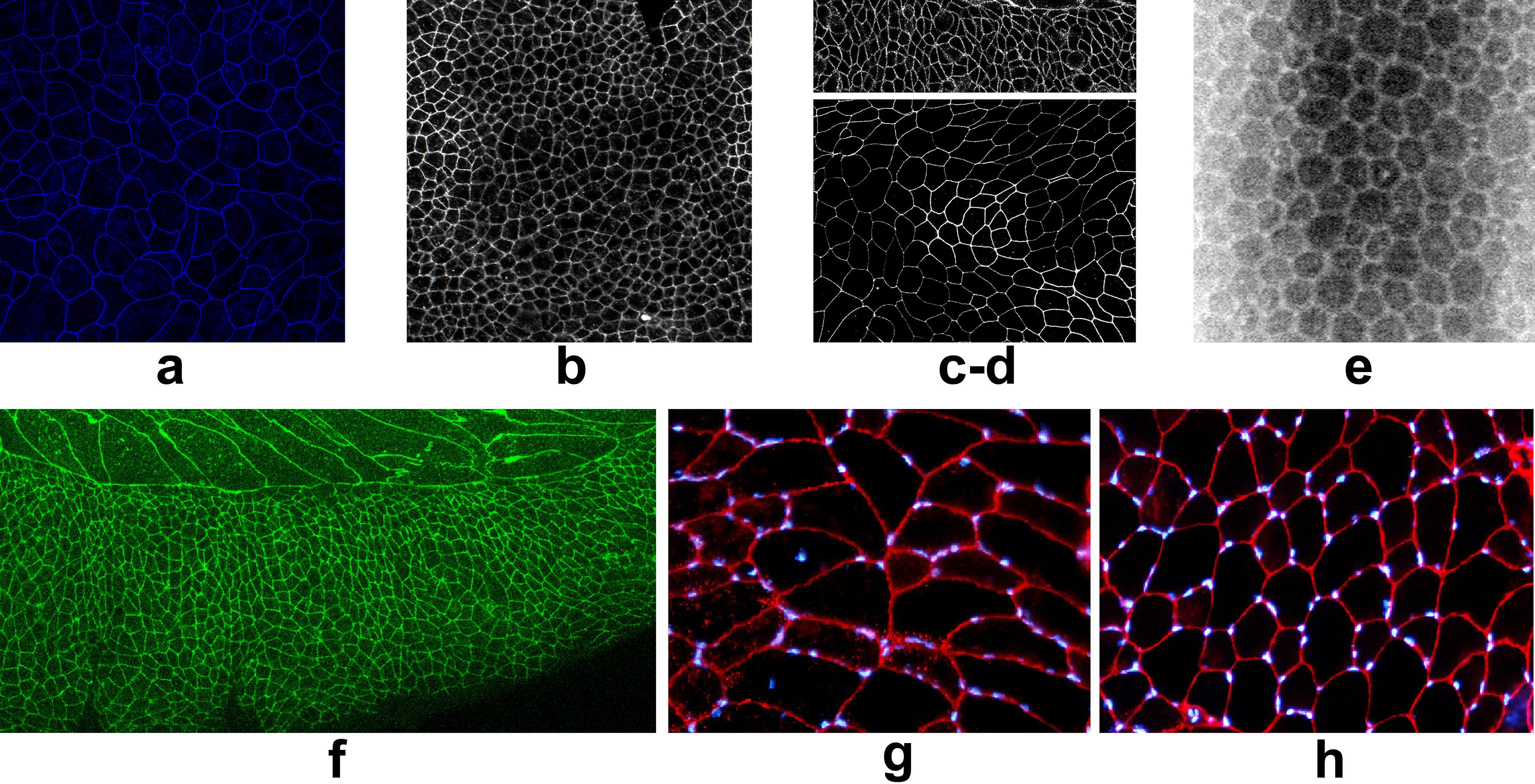
Some examples of the detection and reconstruction procedure are given in the following figures. Tissue images are of different and heterogeneous kinds: drosophila wing epitellium (a-b-c-d-f), cornea (e), muscle fibers (g-h).
A more complete dataset is available here.
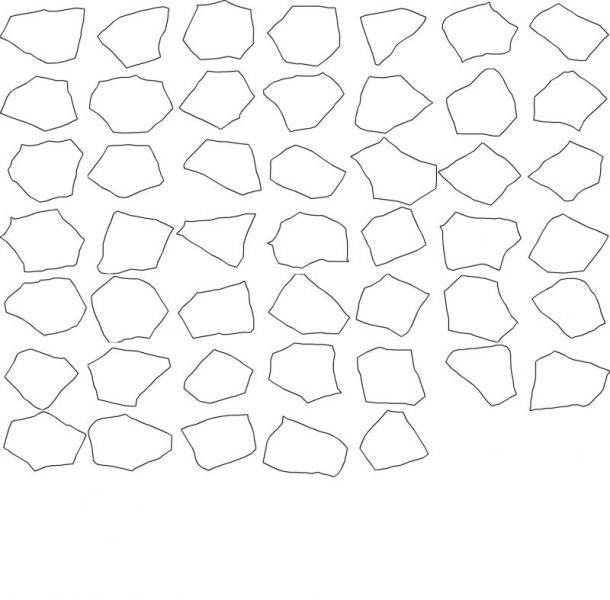
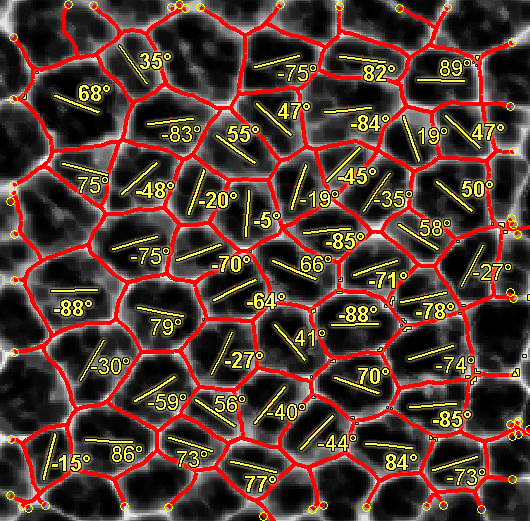
The approach allows for a detailed and exhaustive analysis of the structure morphological features, as for example the precise shape of each sub-structure or the main growth directions.
Collaborations
Prof. Alessandro Abate – Delft Center for Systems and Control, TU Delft, The Netherlands.
Dr. Michela Pozzobon - Istituto di Ricerca Pediatrica - Città della Speranza Foundation, Padova, Italy
Dr. Paolo De Coppi - Great Ormond Street Hospital and Institute of Child Health, University College, London, United Kingdom
References
[1] F. Granera, B. Dolletb, C. Raufastec, and P. Marmottant. Discrete rearranging disordered patterns, part i: robust statistical tools in two or three dimensions. European Physical Journal E, 25:349–369, 2008.
[2] S. Hilgenfeldt, S. Erisken, and R.W. Carthew. Physical modeling of cell geometric order in an epithelial tissue. Proceedings of the National Academy of Sciences, 105(3):907–911, 2008.
[3] D. Ma, K. Amonlirdviman, R. Raffard, A. Abate, C.J. Tomlin, and J.D. Axelrod. Cell packing influences planar cell polarity signaling. The Proceedings of the National Academy of Sciences, 105(48):18800–18805, December 2008.